
STR Profiling
실험실 아카데미
- 세포생물학
- Cell Culture
- Cell Identity
- CO2 Incubator
- Cell Culture 소모품
- 논문
The analysis of Short Tandem Repeats (STR) has become the standard for intra-species identity testing of human cell lines. However, development of markers for authentication of non-human cell lines is still an ongoing process, and more reliable polymeric STR markers which can provide intra-species discrimination have to be identified. To date, a few STR markers have been identified for mouse and African green monkey cell lines [1;2], while others are under development for Chinese hamster and rat cell lines. These are already employed by a small number of companies which also provide STR profiling services for non-human cell lines.
STRs, also called microsatellites, are short tandem repeats of nucleotide sequences consisting of 2‑7 bp, distributed abundantly throughout the genome. 10-15% of the genome encodes genes within the exon regions. In contrast, the enormous remaining nucleotide sequences comprising the introns were thought to be devoid of function and were therefore often referred to as “junk DNA”. However, many specific STRs are located within these introns, and the number of repeats varies between individuals.
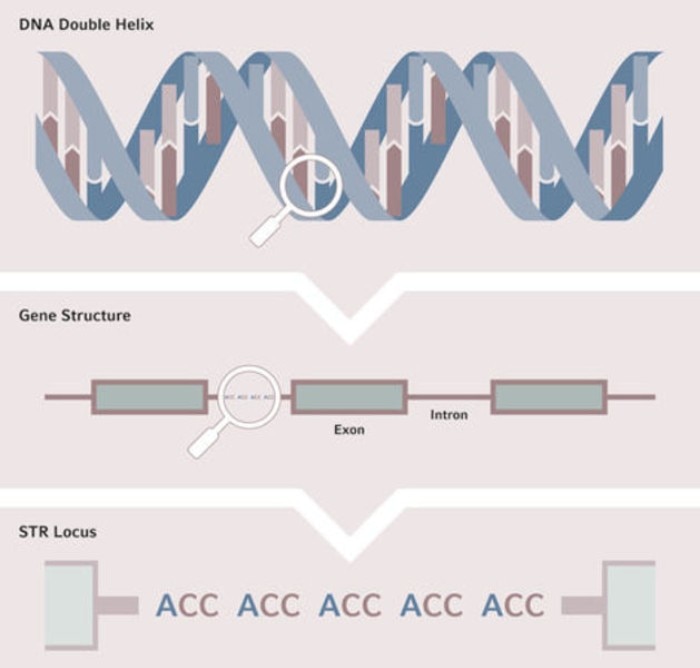
자세히 보기
간단히 보기
The procedure of STR analysis for cell authentication includes several working steps:
자세히 보기
간단히 보기
DNA isolation
For DNA isolation, adherent or suspension cell cultures of interest should be harvested in their exponential growth phase. Some STR profiling service providers also offer to perform DNA isolation from cells, which are spotted on sample collection cards or sent as frozen cell stock or cell pellet.
PCR amplification
During the PCR reaction, STRs are amplified using primer sequences binding to the STR flanking regions. In contrast to the variable STR loci, these flanking regions are highly conserved.
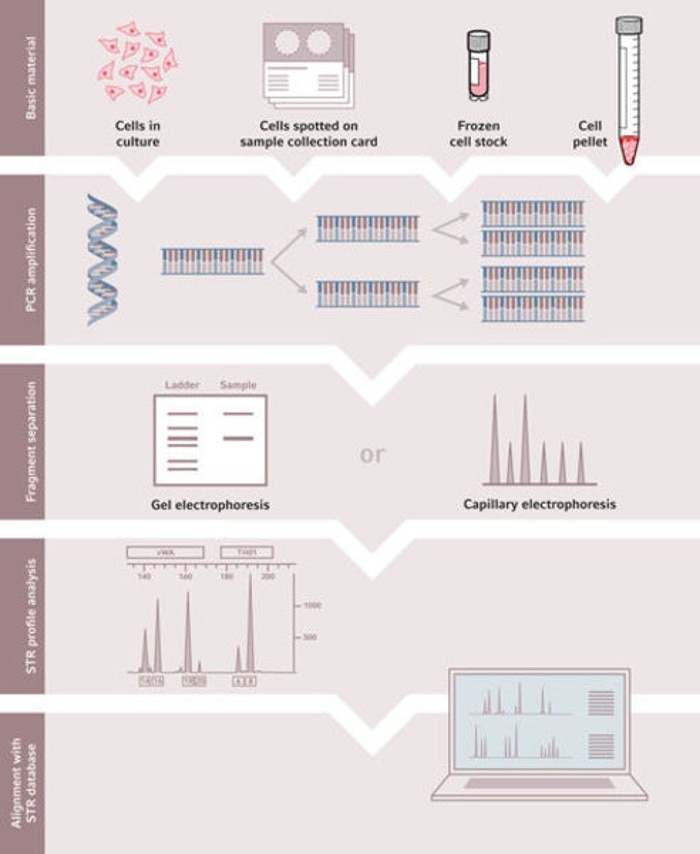
Post-amplification analysis is performed by electrophoresis. Classic gel electrophoresis comprises manual operations. These include gel preparation, loading of samples and gel evaluation by staining procedures or scanning using a gel imager. Capillary electrophoresis (CE) is an alternative to gel-based detection of amplified STR fragments. CE offers high-voltage separation resulting in reduced run times. Combining detection with multi-wavelength analysis, CE instruments can operate with one or multiple capillaries simultaneously, thereby providing a high-throughput method for STR-profiling.
STR profile generation
Genotyping of STR loci is performed by converting the size of amplicons obtained to the number of core sequence repeats using an allelic ladder. The allelic ladder is a marker specifically developed for STR analysis. For each locus analyzed, the number of STR copies for each allele is determined. Thus, an analysis of the recommended 8 STR loci will produce a maximum of 8 numbers if all alleles are homozygous, or 16 numbers if all alleles are heterozygous, respectively. The results can be expressed as an electropherogram or as a text-based table.
Database comparison and cell line authentication
The generated STR profile is compared to an appropriate reference sample. If available, DNA from the donor of the tested cell line is the best comparison. If donor DNA can no longer be accessed, the STR profile obtained for the sample is compared to all STR profiles available using an online database. The databases currently available for comparison are ATCC STR , DSMZ STR, NCBI Biosample, and Cellosaurus. By using a match algorithm, these databases will compare the sample to all available STR profiles and return a list of closest matches. A 100% match is rarely observed. Indeed, cell lines can be unstable, and even with Good Cell Culture Practice, genetic drift is known to occur and generate small variations between cell lines derived from the same donor. To ensure that the tested cell line is reliably identified, a match threshold has to be applied, which is based on the number of shared alleles between two cell lines and expressed as a percentage. According to the consensus standard (ANSI/ATCC ASN-0002-2011) a match between the sample and the reference should be in the 80%-100% range. STR profiles with a percent match between 55% and 80% are probably unrelated. Since this range does include a small number of related cell lines, further data are then needed to confirm or refute any conclusion.
STR profiling can be performed in-house or by a service provider. In-house testing is only recommended if STR profiling is routinely needed and performed according to the consensus standard (ANSI/ATCC ASN-0002-2011). This technique requires a capillary electrophoresis system, which can represent a high financial investment for laboratories. Operating costs as well as personnel training to ensure adequate interpretation of STR profiles must also be considered.
When selecting a STR profiling service provider, three aspects should be taken into consideration: the nature of the sample, the information required, and the final report content. Depending on the service provider, either DNA must be provided by the client directly, or cells may be submitted. If cells are submitted, DNA will then be isolated by the provider.
References:
1. Almeida et al. (2014) Mouse cell line authentication. Cytotechnology. 66(1): 133-47.
2. Almeida et al. (2011) Authentication of African green monkey cell lines using human short tandem repeat markers. BMC biotechnology. 11:102.
3. ANSI/ATCC ASN-0002-2011. Authentication of human cell lines: Standardization of STR profiling.
자세히 보기
간단히 보기
